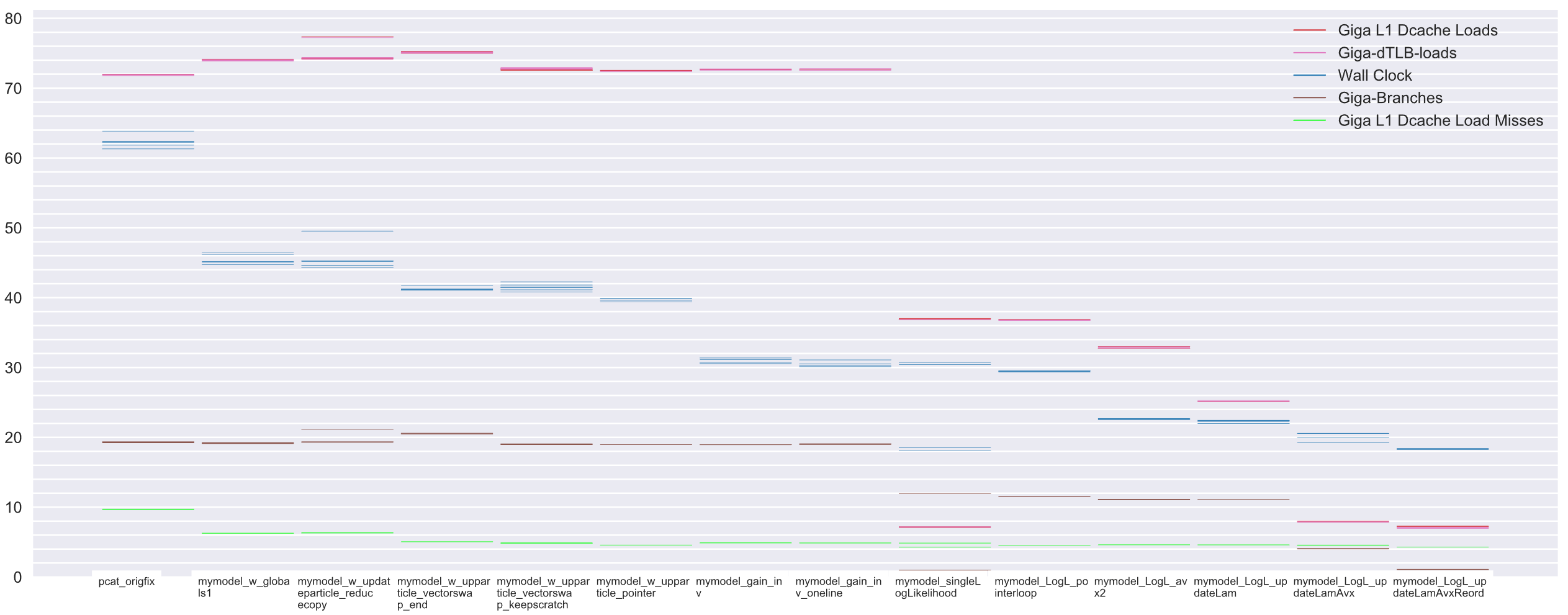
That's the cheatsheet, the operations I performed to get PCat-DNest to run 3.4x its original speed. Each column is a code variation, a small bit faster with each tweak. Some of the gains were expected, such as cutting out big memcopy ops (in the chart, origfix -> mymodel_w_globals1; 27% decrease in runtime!). Others were a surprise, such as removing a divide (mymodel_w_upparticle_pointer -> mymodel_gain_inv; 23.3% runtime reduction). In truth the whole optimization project, the overall gains were a surprise (a pleasant one!). But before I share the tweaks, let me return to the conversation that prompted them.
I heard something unusual the other day. Something strange enough that it snowballed into me doing c++ performance tuning project. A friend of mine was telling me about a scientific research group he worked with. The researchers were writing the new version of their software, he told me, and it would be in python "because the c++ version wasn't fast enough." My ears perked up. The C++ code wasn't "fast" so the response is to abandon the codebase for a language generally considered to be 10x slower?! That idea raised lots and lots of questions. It made no sense!
There was more. I soon came to understand that it wasn't simply that c++ wasn't "fast enough." The situation was that c++ wasn't fast enough to justify the added complexity it introduced. While I could believe C++ added complexity, I had this feeling that something was off with this piece of software. Certainly the solution to "it's slow" is to performance tune the code!
My mind was full of questions. Had the code been tuned? How much performance was being left on the table? How terribly complex was the code? And how much worse would it become if fully tuned? Where did scientific researchers write optimized code, and where did they get into trouble?
I was hooked. I wanted to know how far scientific researchers got before throwing in the towel. The software was open source, and I wanted to know what it could really do. I could have waved my hands around, said something about python being 10x slower, and C++ being a big memory management hassle; estimate some answers to those questions. It wasn't satisfying. I was suddenly wanted to know the performance story for this specific piece of software.
There was only one thing to do. Do a performance analysis and full tuning of the software. So I did. I ended with a pretty great result: 340% speedup and quite a few things learned. Here I will describe 1) performance adjustments I made and their impact 2) performance pitfalls I encountered and tips to avoid 3) discuss some unturned stones, like thread downtime.
The scientific tool in question is PCat-DNest, a statistical astronomy tool I refer to as PCat. PCat takes an astronomical survey image and catalogs the image: finds the location of stars in the image. PCat doesn't find a single set of locations (the highest likelihood catalog). PCat instead samples the distribution of possible star catalogs for the image, and returns a family of possible catalogs. This approach, giving a family of probable answers gives PCat its name: Probabalistic CATaloger.
PCat is built on the statistical sampling framework DNest3. Through DNest3, PCat uses the bayesian technique Markov Chain Monte Carlo (MCMC) using the Metropolis method. Understanding of stars, their density, how they may be modeled using point source function, is used to construct a bayesian. The image data is used to update the bayesian, and PCat jumps around sampling from the universe of all possible star locations to generate a set of samples. Via metropolis, PCat takes its state (catalog of the stars), mutates it slightly, decides if this is a more likely outcome than before, and then either accepts or rejects this "mutant." This mutate-accept/reject processes is repeated, collecting samples.
The processes is similar to an optimizer except that PCat is not targeting a global maximum value and indeed, PCat accepts inferior mutants and walks somewhat randomly.
Importantly for us, this mutate-accept/reject process is the core loop of Pcats and a target for optimization.
- In general, the steps to performance tuning software look something like:
Develop software to minimum/moderate viable functionality
Write tests to guarantee software remains functional as changes made
Create benchmark that will give performance stats
Do performance analysis, make perf changes, check benchmark, repeat, repeat, repeat
Pcat didn't build and didn't have tests (sigh). I hacked away until PCat built, making some minor modifications to library DNest3 the author seems to have made (but didn't document).
Since PCat doesn't have any tests, I made a simple end-to-end test that samples an astro image, and checks the answer against an expected result. PCat is probabilistic which is an issue for reproducibility. I pre-selected the RNG seed, to make my end-to-end test act deterministic. This left limitations -- I had to make certain not to tweak code that affected the order in the random numbers were picked, for example -- but proved effective.
To build my benchmark, I found an image and output catalog settings that took roughly 1min to run. Long enough to average out, but not so long that running several iterations would leave me needing reading material. Variability initially was high, 12% difference between measurements, much too high. I did the following to mitigate:
send all stdio to /dev/null
create a ramdisk to write output data, instead of physical drive
sudo mount -t tmpfs -o size=32m pcat_ramdisk "$RAMDISK_MOUNTPOINT"disable Intel Turbo Boost (allows temporary clock rate boosting as temps allow)
sudo sh -c 'echo 1 > /sys/devices/system/cpu/intel_pstate/no_turbo'
After those changes, variability of wall runtime decreased to 4% in "virgin" PCat; good, but still higher that I would have liked. Honestly, I wasn't thrilled with the measurement inconsistency; mymodel_singleLogLikelihood for example diverged so badly I suspected some gross error in error, but after many attempts I was unable to get more consistent measurements for it. It's a mystery for another day.
I used linux perf-stat to benchmark PCat as it gives gives runtimes and more detailed metrics like L1 loads.
Good. I can build everything in one go, tell what version and commit the binary came from, and extract stats and plots from the benchmark (tooling; I describe those tooling improvements in the appendix, if you're interested). It must be time for code tuning. Let's remove some unnecessary operations and clear up CPU caches! Okay, we need to find a target.
I used Flamegraphs to find and target hotspots. In fact, I created a specific "IPC flamegraph" to help visualize where Instructions Per Cycle (IPC) fall below 1. My graph is quite a bit different from Brandon Gregg's (Flamegraph author) CPI_flamegraph. I wrote a post about it if you're interested.
In addition to flamegraphs I also looked at Intel VTune and while helpful (especially at looking at thread coordination issues), I didn't find it as handy and direct as flamegraphs.
"Vanilla" PCat's flamegraph looks like this:

The red colors indicate where Instructions Per Cycle is <1, and by some, areas where the software is underperforming. Memory operations are slow and are all red, there is no way to avoid that. But functions like pixelLogLikelihood() and logLikelihood() are also red. And similarly, they will always be red because they're floating-point heavy and thus, require multiple cycles per operation. Yes, for PCat, the rule of thumb to "get IPC above 1" is unhelpful and can't be taken literally. If you're doing scientific (FP-math-heavy) computing, you are also likely best to ignore this truism.
Quick note: Click on the flamegraph to open. You can hover over different functions and view quantity and % of cycles for each.
Ignoring the colors, it is abundantly clear from the graph that PCat is spending a lot of time executing __memmove_ operations. In fact, we spend just over 38% of our cycles moving and copying memory. Seems excessive. So that's target number 1: reduce the number of memory operations PCat executes.
The _memmove operations occur within updateParticle(). Looking quickly, this is the core of the MCMC-Metroplis mutate-accept/reject cycle I talked about earlier. Since PCat "walks" through the possibility space, proposing and evaluating mutations, PCat inherently needs to copy some information about the old position in order to create a new proposal. There is no way to get away from a memory operation here, but we can:
Reduce memory copy to minimum necessary data (possibly through use of mask/overlay)
Avoid malloc calls by recyling objects
Find and remove "stupid" memory operations
Starting with reducing data copied. Here in updateParticle() we can see the current position is copied in order to propose a new position.
ModelType proposal = particles[thread][which]; // Did you see the copy? It's right above. // Proposal is ModelType not ModelType& or ModelType *, so that line copies. LikelihoodType logL_proposal = logL[thread][which];
We have to copy the current position to make a proposal, but it would be ideal to copy nothing more than the data needed to uniquely define our position. Poking at the proposal object, MyModel, I found a lot of unneeded fat. Actually, I found a ton of fat. I found that each sample contained an entire copy of the telescope image! The image is shared data, and is certainly not needed to describe a specific set of star positions.
To boost performance, I trimmed down MyModel to a minimum. I stripped out the telescope data and common data and place them in a new object MyModelGlobals, available to all threads by shared pointer. No longer copying full images each cycle, PCat runtime dropped 16.9 seconds, a fantastic 27.1% decrease! 27% for the price of a little object restructuring? Worth it.
(pcat_origfix -> mymodel_w_globals1 in the plot; note L1-dcache-load-misses drop too, a result of not churning duplicate data through the cache)

This alone would have been a nice outcome, but I was suspicous there were other unneeded operations to eliminate. I poked around further and found that indeed, we have other unnecessary copy operations. See if you can spot the issue in updateParticle()?
// Copy the particle ModelType proposal = particles[thread][which]; LikelihoodType logL_proposal = logL[thread][which]; // Perturb the proposal particle double logH = 0.; // Standard Metropolis move logH = proposal.perturb(); ... bool accepted = false; if(levels[thread][indices[thread][which]].get_cutoff() < logL_proposal && randomU() <= exp(logH)) { // Accept particles[thread][which] = proposal; logL[thread][which] = logL_proposal; accepted = true; }
It's in the "Accept" block, the unsuspecting line particles[thread][which] = proposal;.
Proposal is a copy created to be perturbed/mutated. And now that we accept this proposal, we shouldn't need any more copies. We already have an object for the current position and an object for the new position, why would we need a third? The error is the code assigns proposal to a std::vector, and std::vector initiates a copy in order to store the object.
Why does this occur? std::vector needs to store the proposal, but unlike garbage collected (GC) languages like Java, C++ cannot simply hold an object reference and assume the object will persist. No, other code could destroy the object (without a GC or reference counter, C++ leaves the object creator responsible for the lifecyle of the object). So instead, when std::vector receives an object, it stores its own instance of the data, a copy, guaranteeing the data will persist as long as it remains in the vector.
Let's axe this unneeded copy and reclaim some performance! It requires a bit of a hack. Ultimately, the particles must be stored in std::vector, so vector assignment is necessary. Solution is to create the proposal not as a local variable, but as an element within the vector. Below, you can see I create the proposal at the end of the vector.
// Copy the particle - to the end of the list particles[thread].push_back(particles[thread][which]); // Reference for convenient local access ModelType& proposal = particles[thread].back(); logL[thread].push_back(logL[thread][which]); LikelihoodType& logL_proposal = logL[thread].back(); // Perturb the proposal particle double logH = 0.; // Standard Metropolis move logH = proposal.perturb(); ... bool accepted = false; if(levels[thread][indices[thread][which]].get_cutoff() < logL_proposal && randomU() <= exp(logH)) { // Accept std::swap(particles[thread][which], proposal); std::swap(logL[thread][which], logL_proposal); accepted = true; } // Destroy temp particle particles[thread].pop_back(); logL[thread].pop_back();
Essentially, I make std::vector responsible for the persistence of the proposal from the beginning. Makes sense, that's where the object would end up, so it may as well start there. This change drops runtime from 45.5s to 41.3s, a 9.2% reduction. Cool.
(In the chart that's mymodel_w_globals1 -> mymodel_w_upparticle_vectorswap_end)
This optimization suggests another tweak, but the result it turns out, is neutral. You might have noticed that each cycle, I create "scratch" space for the proposal, and then destroy it. Why not keep some persistent scratch? I implemented this, leaving a permanent position at the end of the vector for scratch. This resulted in no change in performance
(Chart mymodel_w_upparticle_vectorswap_end -> mymodel_w_upparticle_vectorswap_keepscratch).
Why? I theorize that the repeated push_back/pop_back operations don't actually trigger a maloc call. At least for primitives, std::vector allocates a contiguous chunk of memory to store data. If you delete just one element, std::vector doesn't free that memory, it just marks that you aren't storing anything there. The same with complex objects I assume. pop_back() seems like it might destroy objects and free memory, but based on std::vector implementation, I don't think it does.
std::vector< std::vector<ModelType> > particles;std::vector< std::vector<ModelType *> > particles;// Copy the particle (to existing scratch object) ModelType *proposal = particles[thread][particles[thread].size() -1]; *proposal = *(particles[thread][which]); LikelihoodType logL_proposal = logL[thread][which]; // Perturb the proposal particle double logH = 0.; // Standard Metropolis move logH = proposal->perturb(); ... bool accepted = false; if(levels[thread][indices[thread][which]].get_cutoff() < logL_proposal && randomU() <= exp(logH)) { // Accept // Stash old particle in scratch slot particles[thread][particles[thread].size() -1] = particles[thread][which]; particles[thread][which] = proposal; logL[thread][which] = logL_proposal; accepted = true; }
This dropped runtime 1.5 seconds, a 3.7% reduction.
(In the chart chart mymodel_w_upparticle_vectorswap_keepscratch -> mymodel_w_uppparticle_pointer).
I'm not exactly sure how to account for this gain. I have a theory that in the std::vector version, std::swap must perform memory operations to move accepted proposal into position. This is based on the assumption that std::vector stores elements in order in a contiguous block of memory, and that swaps require rewriting memory to keep everything contiguous. My pointer version does not care where the particle objects are stored in memory, only that it has an ordered list of their addresses. For the pointers, swapping the position of two elements is a as cheap as swapping 2 pointer values.
If I'm right, that would suggest that for performance, if you have a rapidly changing std::vector of complex objects, it might be better to store pointers in the vector, not the objects themselves. The counterpoint would be that if you need to loop through the vector repeatedly you will incur more pointer chasing, and data locality is lost. How to know? Well, you have to get out those safety goggles and do some benchmarking!
One more thought to finish off memory operations. I've reduced the number of cycles spent on __memmove_avx_unaligned_erms, but not eliminated it. And of course, it would be impossible in MCMC to eliminate all copying. But if we can't can't get rid of __memmove_avx_unaligned_erms.. can we trade it for __memmove_avx_"aligned"_erms. I've heard aligned operations more performant, especially with AVX. Maybe we should align the affected classes!
The "unaligned" part of __memmove_avx_unaligned_erms sounds non-optimal, but it isn't a problem. Glibc has selected this memmove implementation to be the most effective for our system. __memmove_avx_unaligned_erms supports unaligned memory, but this support doesn't mean it is inefficient at moving aligned data. In fact, if we look at glibc, there isn't a memmove_aligned operation. So strike that from your wish list.
But it is neat to see that glibc is using AVX instructions to move our data (AVX2 in our case) allowing 256 bits moves instead of single 64 moves. And from the Glibc source, we can see that if you have a performance/supercomputer x64 chip, Glibc would use AVX-512 and perform 512 bit moves.
- Alright, enough about memory, time for the calculation code. I had a great time spiffing up these sections, and spoiler, ended up rewriting two hotspots using
SIMD. But first, some smaller changes. Sometimes it's the little things in life that make us happy, and in code optimization, sometimes it's the little things that make us fast. Specifically: Transform a divide into a multiply: -23.2% change runtime (!!)
Reorder the
update_lambdasloops: -8.7% change runtime

First the divide. At this point, linux-perf tells us that we spend 51.2% of our cycles in pixelLogLikelihood (flamegraph above shows the same). Looking at the code, I couldn't help noticing that logLikelihood performed a FP divide for every single datapoint. Divides are expensive. An order of magnitude more expensive than a multiply (some resources list FP divide as 20x more expensive than FP multiply). So how could I eliminate the divide? I was lucky, the divide was with a constant, gain.
double variance = (data - bias) / gain; //ignoring dark noise and read noisedouble variance = (data - bias) * gain_inv; //ignoring dark noise and read noiseThat alone dropped runtime from 39.7s to 30.5, a 23.2% reduction!
(In the chart chart mymodel_w_uppparticle_pointer -> mymodel_gain_inv).
Divides, pointer chasing, function calls, these small things really do add up inside hotspots (of course, outside hotspots, such an optimization might just make the code harder to read, with no boost in performance).
Like the logLikelihood calculation, update_lambda loops through all pixels. But further, update_lambda has to pass through each Point Source Founction (PSF, model for a star's radiation), and for each of those, each frequency band, and each template, etc. it's a big 4-nested loop, with different size datasets at different levels.
PCat places at the innermost the template loop, which in my dataset, is a loop of size 2. One level outside this loop, it places the image data loop, which has thousands of iterations. If performance is the name of the game, do we accept this ordering?
x64 has optimizations for simple loops. x64 will detect incremental memory access and prefetch data the loop will need as it cycles. To benefit from these optimizations, we'd like to spend as much time inside long, simple loops as possible. In addition, the CPU has limited on-chip cache, so we want to avoid iterating in such a way that cache is constantly churning (say iterating across datasets that have no shared data between them).
PCat is close, but its loop nesting is not ideal. PCat should place the image data loop as the innermost, so the CPU could zip across a whole image, without worrying about branching, looping through different datasets, wasting its precious cache. Reordering the loops (and precalculating some loop constants) cut 1.75s off runtime, an 8.7% reduction!
(In the chart chart this is the rightmost optimization mymodel_LogL_updateLamAvx -> mymodel_LogL_updateLamAvxReord).

We've realized good performance gains, including a 23.2% runtime decrease just for removing a divide. On the chart, we're in the middle, roughly mymodel_LogL_pointerloop. That blue line has dropped a lot! Still, two big hotspots that still remain: logLikelihood() and update_lambdas() and together they consume 67% of all cycles. Well, these functions are just loops that repeatedly perform the same math operations, why not use SIMD?
Single Instruction, Multiple Data (SIMD) instructions, are a performance tool. x64 has hardware optimizations that allow it to rapidly, say, perform addition on 4 doubles at the same time. It costs more than an individual addition, but not as much as 4 individual additions. For x64, the widest, most commonly available SIMD instructions are AVX2. Most people have heard of SSE, but SSE4 gives only 128 bit registers (2 double operations) while AVX2 allows for 256 bit operations (4 doubles). Maybe you're lucky enough to have an Intel "high performance" chip (server and supercomputer chips), in which case you have AVX-512 and can processes 8 doubles (512 bits) simultaneously. AVX2 is extremely common while AVX-512 is rare. Thus I targeted AVX2 for my SIMD optimizations.
If my chip has these performance SIMD instructions, you might ask, why doesn't my compiler make use of them? Indeed, in some cases the complier will use SIMD instructions if it is confident the math operations are truly parallel and independent. I checked my PCat binary but found that gcc had not used SIMD instructions, possibly because logLikelihood() math is a set of calculations with several stages and references to instance variables. I suspect the compiler cannot be certain each loop is performing the same, independent calculation.
If you find, that your compiler didn't give you the SIMD instructions you wanted, you have two options. 1) You can write some assembly, or 2) you can use compiler intrinsics. You want to use intrinsics. They're functions that look like SSE/AVX instructions, but they return "variables" and can refer to variables, which means the compiler, not you, gets to worry about which exact registers to use for the computation. Intrinsics are really awesome!
Original (with some pointerification) logLikelihood() looks like
double SloanModel::logLikelihood() const { double logL = 0.; double *dataPos = &globals->data[0]; double *dataLast = &globals->data[nbin*npsf*npix]; double *lambdaPos = &this->lambda[0]; for (; dataPos<=dataLast; dataPos++, lambdaPos++){ logL += std::pow(*lambdaPos - *dataPos, 2) / (-2 * gain_inv * (*dataPos - bias)); } return logL; }
Adding the instrinsics, the beginning of the loop looks like:
double *dataPos = &globals->data[0]; double *dataLast = &globals->data[nbin * npsf * npix -1]; double *dataLastChunk = (double *) ((uintptr_t) (dataLast - 3) & ~0b11111); double *lambdaPos = &this->lambda[0]; // Init 4-wide constants __m256d gain_pd = _mm256_set1_pd(gain_inv); __m256d neg2_pd = _mm256_set1_pd(-2); __m256d bias_pd = _mm256_set1_pd(bias); __m128d logL_sd = _mm_setzero_pd(); for (;dataPos <= dataLastChunk;) { __m256d _data = _mm256_load_pd(dataPos); __m256d numerator = _mm256_sub_pd(_mm256_load_pd(lambdaPos), _data); numerator = _mm256_mul_pd(numerator, numerator); __m256d denominator = _mm256_mul_pd(_mm256_mul_pd(neg2_pd, gain_pd), _mm256_sub_pd(_data, bias_pd)); numerator = _mm256_div_pd(numerator, denominator); // Sum 4 results horizontally into logL_sd numerator = _mm256_hadd_pd(numerator, numerator); __m128d tmp = _mm256_extractf128_pd(numerator, 1); tmp = _mm_add_sd(_mm256_castpd256_pd128(numerator), tmp); logL_sd = _mm_add_sd(logL_sd, tmp); dataPos += 4; lambdaPos += 4; } ...
double *dataLastChunk = (double *) ((uintptr_t) (dataLast - 3) & ~0b11111);
Is some bookkeeping to find the find the last fully-filled, 4-double, 32-bytes address. I pull down doubles in sets of 4, so without the bookkeeping, I could easily read 32 bytes of memory where the image only has, say, 16 bytes remaining, resulting in a memory bounds violation, and an undesired segfault. Instead I safely process as many 4-double chunks as are available, and then use a "peel" loop to process the remaining 1-3 values (peel loop is basically the same as the original loop).
Looking at intrinsics in action, instead of writing out a vmovapd instruction to load our data into the AVX register, I write _mm256_load_pd(dataPos). That gives a nice, typed __m256d back, and the compiler will figure out what registers to use. Same for the math, instead of vmulpd, it's _mm256_mul_pd; instead of vsubpd, it's _mm256_sub_pd, in all cases returning results as a "variable."
Overal, the intrinsics are pretty straightforward, except for one thing: summing several doubles together "horizontally." I have 4 doubles packed into numerator that need to get summed with each other and with logL_sd. This requires first, summing pairs of doubles (_mm256_hadd_pd) removing the high order value (_mm256_extractf128_pd) and summing again (_mm_add_sd). The instructions themselves are responsible for the confusing operation (there is no sum-4-horizontal-doubles instruction), the intrinsics do not provide operations not found in an instruction. Written in raw assembly, this sum would require just as many steps.
AVX2 SIMD give us?LogLikelihood() converted to AVX2 went from 29.4s to 22.6s runtime, a whopping 6.83s saving (23.2%).updateLambda() to AVX2 intrinsics took runtime from 22.6s to 20.1s, a 11.3% reduction. Combined with the loop reordering I wrote about above, that dropped further to 18.3s.With both LogLikelihood() and updateLambda() converted to AVX2 intrinsics, I've dropped runtime 37.8%. That's a real nice gain! Outside of program hotspots, the hassle of adding the SIMD intrinsics is probably not worth it, but in hotspots where it will shave 37.8% off the runtime, it certainly is.
There is certainly more performance that can be wrung out of PCat. As a start, PCat repeatedly executes the metropolis propose-test-accept/reject cycle, copying state data around. I suspect that data can be trimmed further into an even more minimal proposal object than I was able to acheive. Or a step further, it would be faster to not perform the copy operation at all. It might be possible to treat the proposal as a mask or an overlay on an existing object. This would incur a follow-mask step in image calculations, so there is risk this added work would negate the savings of having the mask/overlay, but it is a possability.
On tools, other c++ compilers promise better performance than gcc. Some involve benchmarking the program and performing a second pass compile that attempts to optimize hotspots. At the risk of having to fuss with compiler flags and linking, using another compiler could be a worthwhile experiment.
Optimize data layout, for better cache utilization. update_lambdas() remains as a huge hotspot, and a complex 4-nested loop. This 4-nested loop might benefit from storing data such that values have good locality with other data used in computation. I suspect caching can be optimized, cache line efficiency improved.
Optimize thread synchronization. A quick analysis with Intel VTune shows that PCat-Dnest's threads pause in order to synchronize and also to write data to stdout and disk. I'm certain these pauses could be reduced. Do the threads truly need to halt and synchronize? If the threads simply need to make decisions or set parameters, couldn't the first thread past the barrier leave parameters for the trailing threads, and keep on computing? Further, PCat threads could spend more time computing if stdout and disk output was handled by writer threads.
So, after performing a lot of benchmarks, looking at flamegraphs, tweaking hotspots, and writing some SIMD code, I managed a significant increase in performance. PCat's speed is now 3.4x what it used to be, that means the original benchmark that took 62.3s now takes only 18.3s! In the chart, the blue line is down significantly.

So what of the astronomy researchers and their plan to write the next version in Python? Well, they didn't performance tune their code, and this lost them 3.4x speedup.. though tuning does require work (writing SIMD intrinsics for example). I managed 1.9x speedup without SIMD, so let's say that the researchers left 1.9x on the table. And let's say that the Python version is 3x slower than the non-optimized c++ codebase (I think this is a reasonable estimate assuming they keep calculations within Numpy). That leaves the Python version 6x slower than my optimized version (10x if you include SIMD). Thats a lot of performance to leave behind.
No I think the astronomy researchers were interested in performance, but it was not a true requirement for the project. PCat is a proof of concept. It functions, but it is very rough and would require a fair amount of work to be turned into a solid, ready-for-astronomy tool. Writing a proof of concept, it does seem the flexibility and convenience of Python are a good fit. Proof of concepts don't need to be high performance.
If you happen to be in PCat's shoes, and have a C++ scientific computing project that doesn't perform, here's my executive summary of what to do:
- Basic tips:
Compile using -O3, --ffast-math, and other optimizations (where compatible)
- Understand C++ pass-by-value, be careful to not trigger excess memcpy ops
If using vectors, keep objects in-vector and use references for local vars
Profile code (suggest flamegraphs) so you know your hotspots
- Reduce memory allocation and copies (expensive)
Include "scratch space" objects for computation instead of creating/destroying repeatedly
If copying is necessary, keep copied objects small
Structure nested loops to have long stretches of computation instead of stutter segments
- Avoid repeated divides
Consider using log-subtraction instead
- More time-intensive tips:
Consider Intel
MKLif using advanced math calculations- Write benchmarks and tests (at least simple end-to-end tests)
Be confident performance tweaks aren't changing behavior of program
Know if performance tweaks make a difference
- Profile and optimize hotspots
Use
flamegraphsbased on cpu-cycle samplingRemove unneeded
memcopy/movesOptimize math, consider log-subtraction/addition instead of divides/multiplies
Optimize loops, data locality
- Structure computation to avoid blocking on io (use writer threads)
Even small stdio/disk writes can cause serious blocking
- Consider rewriting hotspots with SIMD
Pick widest:
AVX-512,AVX2,SSE4in that order based on processor availabilityUse compiler intrinsics
Consider alternate/optimizing compiliers for further speedup
Finally, performance analysis is a necessary stage of software development (assuming you care an about performance). Software doesn't become fast by being written in C++. Software becomes fast by being performance tuned. Setting up instrumentation, running benchmarks, making repeated small tweaks, it seems like a chore but, 1) it provides results, 2) I find it's a pretty interesting chore. It's interesting to discover where in the code the hotspots are, see what code changes reduce runtime, and to write SIMD intrinsics.
In the adage "Premature optimization is the root of all evil," premature might be the root of evil, but certainly "failure to optimize" is at least a serious sin. Don't sin, speed instead ;-)
For fun, the initial flamegraph:

The final flamegraph:

As mentioned above, I made a few tooling improvements beyond just getting the end-to-end benchmark running. I find the return on good, minimal tools to be excellent (and they make work much more pleasant!). To be able to benchmark more rapidly, with better clarity, and useful numbers, I did the following:
Rewrite the build system in cmake. I included PCat's 2 dependencies as cmake libraries so I could make changes in the libraries, and quickly rebuild the whole enchilada. This was very useful when I made repeated tweaks to lib DNest3.
Modify PCat and library to include build information with the binary.
I was about to run a lot of benchmarks with slighly different versions of PCat. I wanted to make it easy to grab the build flags and git commit for any build, to double check I was benchmarking what I thought I was benchmarking. For completeless, PCat included full build info for its dependencies, as well as itself.
In cmake this looked like:
file(WRITE ${CMAKE_BINARY_DIR}/buildinfo.h "constexpr char BUILDINFO[] = R\"(${CONTENTS})\";\n") include_directories(${CMAKE_BINARY_DIR})
Where CONTENTS was a string containing $CMAKE_CXX_FLAGS and the latest git commit info.
Back at the c++ codebase, main() simply included a check for arg --info, and output BUILDINFO[] when the flag is found.
With this the benchmark could automatically exec and log --info for reference (of course, us humans would never get confused about which commit each of the 16 binaries came from, --info is just for the automation ;-] )
$ ./pcat --info #### PCAT-DNEST FLAGS: BUILD_ARCH Linux-5.0.0-31-generic BUILD_TYPE release CXX_FLAGS -Wall -Wextra -pedantic -Wno-unused-parameter CXX_FLAGS_DEBUG -O0 -DNDEBUG -g -fno-omit-frame-pointer CXX_FLAGS_RELEASE -O3 -DNDEBUG -g -funroll-loops -fno-omit-frame-pointer BUILD_CMAKE_VER 3.10.2 BUILD_COMP x86_64 ... #### LIB RJObject FLAGS: BUILD_ARCH Linux-5.0.0-31-generic BUILD_TYPE release CXX_FLAGS -Wall -Wextra -pedantic -ansi CXX_FLAGS_DEBUG -O0 -g -fno-omit-frame-pointer CXX_FLAGS_RELEASE -O3 -flto -march=native -g -fno-omit-frame-pointer BUILD_CMAKE_VER 3.10.2 BUILD_COMP x86_64 ### SRC LAST COMMIT bf28695 - (HEAD -> mymodel_loglikely_pointer_n_simd) Reorder updatelambdas to give avx longer streams of data. <Colin> ###
Write helper scripts to extract values from the perf-stat output, plot the data using matplotlib, and write csv.
Mean values would have been enough, but looking at each builds runtime side-by-side gives a better understanding. You can get an impression of magnitude, and of which metrics the build affected. Look at the plot, see if you think you could get as good an understanding without a visual representation. (BTW, plots in this post were all generated by my benchmarks).